Archive for category data standards
DITA – A framework for scientific publishing?
Posted by peanutbutter in data standards, Journals Publishing on February 1, 2011
There are two industry recognised standards for XML based documentation. These are Docbook and DITA (Darwin Information Typing Architecture).
Docbook is the older of the two specifications and created specifically for technical documentation. DITA, is a younger specification which grew out of IBM, and is referred to as having its own architecture and was designed to provide structure to more than just a book. Both specifications are OASIS standards.
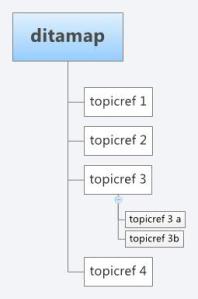
As with XML schemas, both specifications can be extended to include bespoke features. However, Docbook is based more on a book structure with Sections and subsections, where as DITA is built around topics that can be built up in any arrangement based on a document map. A DITA topic is open to specialisation itself, however, a topic has only three required elements
- An id attribute
- A title
- A body
- ,
A topic can exist as a single XML file which can be composed into any arrangement for publication through the use of a document map. A DITA structure would present a more flexible architecture where the same “topic”, i.e a journal article section, such as an abstract, materials and methods, or results, could be included with ease more than one publication, correctly referenced. In this respect DITA is more like an object-oriented document schema, and can be more easily repurposed (in terms of structure) for any output format (i.e pdf, HTML). In the same respect, Docbook can be configured with some work to behave on a more topic by topic basis and DITA can support a book based methodology. They are after all both XML schemas and are equally extensible or open to specialisation.
As its a standard, whole ecosystems have emerged which makes use of the DITA architecture. For example, DITA for publishers provides libraries to convert DITA markup into HTML, PDF, EPUB, and Kindle rendering support. This allows content structures in DITA to be repurposed for different audiences or different devices with relative ease.
I have recently started using DITA as an architecture to represent content, primarily designed for books. However, with new demands appearing for different delivery mechanisms of the traditional textbook, such as Web delivery and ebooks, DITA is proving to be immensely powerful to deliver the same content through different mediums with relative ease and speed. In using it, it seems obvious that a DITA architecture would benefit the representation of content within a journal article, allowing references re-purposing and multiple format delivery. Maybe a topic for discussion through the Beyond the PDF forum.
In the end, it’s just XML, so I wont repeat the virtues of content markup through XML. However, for me its main advantage is the object oriented -like topic structure as a working architecture.
Related articles
- Using RDFa with DITA and DocBook (devx.com)
- Dita Educational Use cases (docs.oasis-open.org)
- Converting documents between a wiki and Word, XML, FrameMaker or other help formats (ffeathers.wordpress.com)
- Future-proofing e-books with XML (teleread.com)
- The PDF Landscape for DITA Content (tc.eserver.org)
HUPO PSI-PAR: standard format for protein affinity reagents
Posted by peanutbutter in data standards on April 7, 2009

- Image via Wikipedia
HUPO PSI-PAR: standard format for protein affinity reagents is now available for Public Comment on the PSI Web site for the next 30 days. The public comment period enables the wider community to provide feedback on a proposed standard before it is formally accepted, and thus is an important step in the standardisation process. This message is to encourage you to contribute to the standards development activity by commenting on the material that is available online. We invite both positive and negative comments. If negative comments are being made, these could be on the relevance, clarity, correctness, appropriateness, etc, of the proposal as a whole or of specific parts of the proposal. If you do not feel well placed to comment on this document, but know someone who may be, please consider forwarding this request. There is no requirement that people commenting should have had any prior contact with the PSI. If you have comments that you would like to make but would prefer not to make public, please email the PSI-Editor directly.
The Triumvirate of Scientific Data
Posted by peanutbutter in bioinformatics, data standards, ontology, open data on October 30, 2008
 |
In a recent Nature editorial entitled Standardizing data, several projects were highlighted that are forfeiting there chances of winning a Nobel prize (according to Quackenbush) and championing the blue collar science of data standardization.in the life-sciences.
I wanted to take the article a step further highlight three significant properties of scientific data that I believe to be fundamental in considering how to curate, standardize or simply represent scientific data; from primary data, to lab books, to publication. These significant properties of scientific data are the content, syntax, and semantics, or more simply put -What do we want to say? How do we say it? What does it all mean? These three significant properties of data are what I refer to as the Triumvirate of scientific data.
Content: What do we want to say?
Data Content is defined as the items, topics or information that is “contained in” or represented by a data object. What is, should or must be said. Generic data content standards exists, such as Dublin Core, as well as more focused or domain specific standards. Most aspects of the research life-cycle have a content standard. For example, when submitting a manuscript to a scientific publisher you are required to conform to a content standard for that Journal. For example, PlosOne calls their content standard Criteria for Publication and lists seven points to conform to.
The Minimum Information about [insert favourite technology] are efforts by the relevant communities to define content standards for their experiments. These do (should) not define how the content is represented (in a database or file format) rather they state what information is required to describe an experiment. Collecting and defining content standards for the life-sciences is the premise of the MIBBI project.
Syntax: How do we say it?
The content of data is independent of any structure, language implementation or semantics. For example when viewing a journal article on Biomed central you typically have the option to view or download the “Full Text” which is often represented in HTML or you have the option of viewing the PDF file or XML. Each representation has the same scientific content to a human but is structured and then rendered (or “presented”) to the user in three different syntax.
The majority of the structural of syntactic representation of scientific data is largely database centric. However, alternative methods can be identified such as Wikis (OpenWetWare, UsefulChem), Blogs (LaBLog), XML, (GelML), RDF (UniProt export) or described as a data model (FuGE) which can be realised in multiple syntax
Semantics: What do we mean?
The explicit meaning of data is very difficult to get right and is a difficult problem in the life-sciences. One word can have many meanings and one meaning can be described by many words. A good example of a failure to correctly determine the semantics of data is described in the paper by Zeeberg et al 2004. In the paper they describe the mis-interpretation of the semantics of gene names. This mis-interpretation of semantics resulted in an irreversible conversion to date-format by Excel and which percolated through to the curated LocusLink public repository.
Within the life-sciences the issue of semantics is being addressed via the use of Controlled vocabularies and ontologies.
According to the Neurocommons definition; A controlled vocabulary is an association between formal names (identifiers) and their definitions. A ontology is a controlled vocabulary augmented with logical constraints that describe their interrelationships. Not only do we need semantics for data, we need shared semantics, so that we are able to describe data consistently, within laboratories, across collaborations and transcending scientific domains. The OBO Foundry is one of the projects tasked with fostering the orthogonal development of ontologies – one term only appears in one ontology and is referenced by others – with the goal of shared semantics.
Summary
When considering how to curate, standardize or represent scientific data, either internally within laboratories, or externally for publication, the three significant properties of content, syntax and semantics should be considered carefully for the specific data. Consistent representation of data conforming to the Triumvirate of scientific data will provide a platform for the dissemination, interpretation, evaluation and advancement of scientific knowledge.
Acknowledgments
Thanks to Phil Lord for helpful discussions on the Triumvirate of data
Conflict of interest
I am involved in the MIBBI project, the development of GelML and a member of the OBO Foundry via the OBI project.
MIAPE: Gel Informatics is now available for Public Comment
Posted by peanutbutter in bioinformatics, data standards, Journals Publishing, MIBBI, Proteomics on August 18, 2008

PSI logo
The MIAPE: Gel Informatics module formalised by the Proteomics Standards Initiative (PSI) now available for Public Comment on the PSI Web site. Typically alot of this information will be contained in the image analysis software, so we would especially encourage software vendors to review the document. The public
comment period enables the wider community to provide feedback on a proposed standard before it is formally accepted, and thus is an important step in the standardisation process.
This message is to encourage you to contribute to the standards development activity by commenting on the material that is available online. We invite both positive and negative comments. If negative comments are being made, these could be on the relevance, clarity, correctness, appropriateness, etc, of the proposal as a whole or of specific parts of the proposal.
If you do not feel well placed to comment on this document, but know someone who may be, please consider forwarding this request. There is no requirement that people commenting should have had any prior contact with the PSI.
If you have comments that you would like to make but would prefer not to make public, please email the PSI editor Norman Paton.
PEFF:A Common Sequence Database Format in Proteomics
Posted by peanutbutter in data standards, development on August 13, 2008
 Image via Wikipedia
Image via Wikipedia PEFF:A Common Sequence Database Format in Proteomics is now available for Public Comment on the PSI Web site (http://psidev.info/index.php?q=node/363). The public comment period enables the wider community to provide feedback on a proposed standard before it is formally accepted, and thus is an important step in the standardisation process.
This document presents a unified format for protein and nucleotide sequence databases to be used by sequence search engines and other associated tools (spectra library search tools, sequence alignment software, data repositories, etc). This format enables consistent extraction, display and processing of information such as protein/nucleotide sequence database entry identifier, description, taxonomy, etc. across software platforms. It also allows the representation of structural annotations such as post-translational modifications, mutations and other processing events. The proposed format has the form of a flat file that extends the formalism of the individual sequence entries as presented in a FASTA format and that includes a header of meta data to describe relevant information about the database(s) from which the sequence has been obtained (i.e., name, version, etc). The format is named PEFF (PSI Extended FASTA Format). Sequence database providers are encouraged to generate this format as part of their release policy or to provide appropriate converters that can be incorporated into processing tools.
This is an announcement to encourage you to contribute to the standards development activity by commenting on the material that is available online. We invite both positive and negative comments. If negative comments are being made, these could be on the relevance, clarity, correctness, appropriateness, etc, of the proposal as a whole or of specific parts of the proposal.
If you do not feel well placed to comment on this document, but know someone who may be, please consider alerting them towards this information. There is no requirement that people commenting should have had any prior contact with the PSI
Double standards in Nature biotechnology
Posted by peanutbutter in bioinformatics, data standards, Journals Publishing, MIBBI on August 7, 2008
OK, So that is a relatively inflammatory and controversial headline, edging on the side of tabloid sensationalism. What is refers to is probably a situation that I may never find myself in again, which is in this months edition of Nature Biotechnology I am an author on two, biological standards related publications.
The first is a letter advertising the PSI’s MIAPE Guidelines for reporting the use of gel electrophoresis in proteomics. This letter is also accompanied by letters referring to the MIAPE guidelines for Mass Spectrometry, Mass Spectrometry Informatics and protein modification data.
The second is a paper on the Minimum Information about a Biomedical or Biological Investigations (MIBBI) registry entitled Promoting coherent minimum reporting guidelines for biological and biomedical investigations: the MIBBI project.
The following press release describes this paper in more detail.
More than 20 grass-roots standardisation groups, led by scientists at the European Bioinformatics Institute (EMBL-EBI) and the Centre for Ecology & Hydrology (CEH), have combined forces to form the “Minimum Information about a Biomedical or Biological Investigation” (MIBBI) initiative. Their aim is to harmonise standards for high-throughput biology, and their methodology is described in a Commentary article, published today in the journal Nature Biotechnology.
Data standards are increasingly vital to scientific progress, as groups from around the world look to share their data and mine it more effectively. But the proliferation of projects to build “Minimum Information” checklists that describe experimental procedures was beginning to create problems. “There was no way of even finding all the current checklist projects without days of googling,” says the EMBL-EBI’s Chris Taylor, who shares first authorship of the paper with Dawn Field (CEH) and Susanna-Assunta Sansone (EMBL-EBI). “As a result, much of the great work that’s going into developing community standards was being overlooked, and different communities were at risk of developing mutually incompatible standards. MIBBI will help to prevent them from reinventing the wheel.
The MIBBI Portal already offers a one-stop shop for researchers, funders, journals and reviewers searching for a comprehensive list of minimum information checklists. The next step will be to build the MIBBI Foundry, which will bring together diverse communities to rationalise and streamline standardisation efforts. “Communities working together through MIBBI will produce non-overlapping minimal information modules,” says CEH’s Dawn Field. “The idea is that each checklist will fit neatly into a jigsaw, with each community being able to take the pieces that are relevant to them.” Some, such as checklists describing the nature of a biological sample used for an experiment, will be relevant to many communities, whereas others, such as standards for describing a flow cytometry experiment, may be developed and used by a subset of communities.
“MIBBI represents the first new effort taking the Open Biomedical Ontologies (OBO) as its role model”, says Susanna-Assunta Sansone. “The MIBBI Portal operates in a manner analogous to OBO as an open information resource, while the MIBBI Foundry fosters collaborative development and integration of checklists into self-contained modules just like the OBO Foundry does for the ontologies”.
There is a growing understanding of the value of such minimal information standards among biologists and an increased willingness to work together across disciplinary boundaries. The benefits include making experimental data more reproducible and allowing more powerful analyses over diverse sets of data. New checklist communities are encouraged to register with MIBBI and consider joining the MIBBI Foundry.
Press release issued by the EMBL-European Bioinformatics Institute and the Centre for Ecology and Hydrology, UK.
CARMEN – A Scalable Science cloud
Posted by peanutbutter in bioinformatics, CARMEN, cloud, conference, data standards, neuroinformatics, open science, video on June 25, 2008
Paul Watson presents a talk on CARMEN a the Google Seattle Conference on Scalability.
Proteomics Standards Initiative recommendations
Posted by peanutbutter in bioinformatics, data standards, development, Proteomics, Uncategorized on June 12, 2008
Several new standard artefacts have progressed through the public consultation process of the PSI. They are the MIAPE Column Chromatography document and the Mass Spectrometer Markup Language Specification (mzML).



![Reblog this post [with Zemanta]](https://i0.wp.com/img.zemanta.com/reblog_e.png)